JSTOR generative AI pilot - Or is Semantic Search coming for academic databases?
A decade ago in 2012, I observed how the dominance of Google had slowly affected how Academic databases and OPACs/ catalogues (now discovery services) work.
In a nutshell I argued that due to Google's influence, academic search at the time had already moved towards ranking results by relevancy by default (as opposed to sorting by date or accession number), adopting implicit AND (as opposed to requiring strict AND) and was slowly moving toward auto- stemming by default and searching over full-text (as opposed to just abstracts, keywords).
This has mostly happened. But I also predicted
But unless a miracle happens, will NEVER - Do a "soft AND", where occasionally search terms might be dropped
Here what I was trying to say is that there is little chance of what is often called keyword or lexical based + strict or mostly strict Boolean search (typically implemented with BM25/TF-IDF) going away.
The alternative at the time was non-boolean search algos but that were mostly still keyword or lexical based (e.g. non-boolean vector space search), but these days we are talking about semantic search which obviously isn't boolean.
However, I suspect things are now changing and this blog post I will explain why. I will also do a brief overview of JSTOR's generative AI pilot as of Nov 2023 where they introduced an experimental search that "understands your query and provides more relevant results, even if you don't use the exact words.", clearly a semantic search as an alternative to the traditional search labelled "keyword-based results"
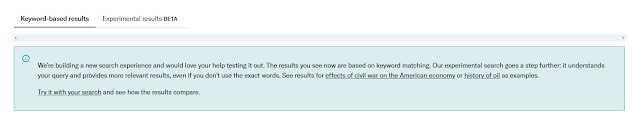
My initial findings are that the new experimental semantic search relevancy rankings seem to be really good, outperforming the traditional status quo keyword search most of the time. There might even be some indications the results are even better if you search in natural language style than keyword style!
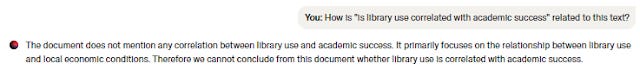
JSTOR Pilot also introduces some novel ideas on how to mitigate the downsides of the unpredictability of Semantic Search by leveraging language models to ask "how is <query> related to this text" over the full-text!
I end with a discussion about the irony of how LLM powered search tools like elicit.com, scispace might be bringing us back to the old days of Dialog where searches were charged per use/time and there was a need to be efficient with nested Boolean strategies. Except these are also the tools most likely not to support Boolean!
Introduction
In my talks about generative AI/Large Language models, I have this standard slide where I assert the three typical ways academic search engines are incorporating the benefits of LLMs.

Of these three benefits, the middle one - Generated of Direct answers with citations (using RAG or retrieval augmented generation) is the one that gets all the attention because it is the most eye-catching feature and promises to disrupt the decades old paradigm of showing you N top documents that might answer your query.
The third benefit, extraction of information from papers can also affect relevancy if it is used pre-search for subject or term extraction. For example, PubMed seems to be doing auto-indexing of MeSH headings since April 2022, and the inaccuracy of the extraction might be affecting reliability of search queries!
Vendors seem to agree, and pretty much everyone from Dimensions to Scopus AI to experiments by Exlibris all seem to be rushing to add this feature at least.
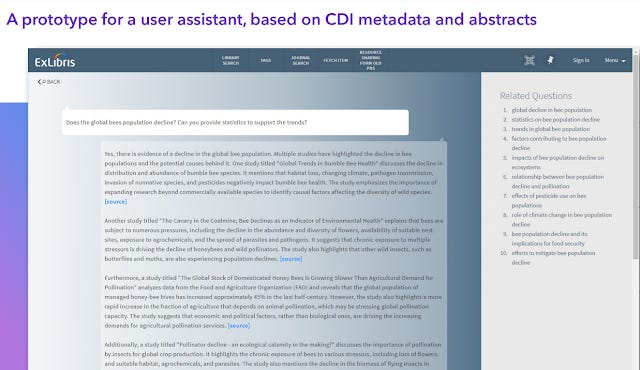
Exlibris prototype search that generates a direct answer using abstracts
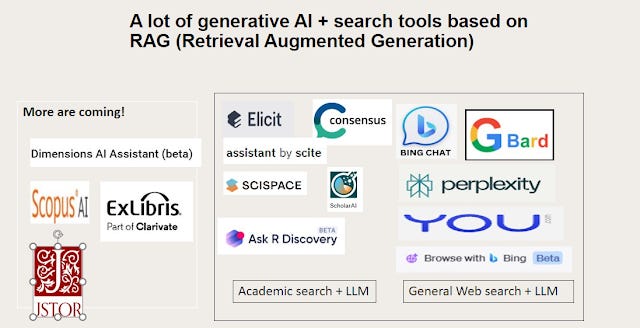
See my updated list of Academic Search engines using Retrieval Augmented Generation
It's less clear how many academic search engines are moving away from traditional keyword based searches towards more semantic type searches using embedding though the fact that many of them work well even if you type with long natural language queries is suggestive of what is happening (though not 100% conclusive).
Also just because a search system can generate a direct answer does not imply it is not using a keyword-based system to find documents. You could in theory find relevant docs using Boolean+TF-IDF for the retriever part of RAG. That said many such systems are already heavily using large language models for generation and summarization/extraction, it seems unlikely they are just using traditional TF-IDF, BM25 only methods (also known as sparse embeddings) as opposed to dense embeddings of some kind (eg. BERT, MiniLM-L6-v2,text-embedding-ada-002 etc)
The line between keyword based (typically implemented by TF-IDF/BM25) and semantic search (these days using some form of embeddings) can be a bit difficult to define so below I try to entangle this.
Experts in information retrieval can skip this long section to my overview of JSTOR generative AI pilot.
What do we mean by keyword-based or lexical search vs Semantic search?
Post edit note: I took a long time to write this long section but am still not totally happy with it. I later wrote on Medium another piece - Boolean vs Keyword/Lexical search vs Semantic — keeping things straight which I am happier with articulating the differences between Boolean, Keyword/lexical search and Semantic Search.
The Boolean model of information retrieval (BIR) was proposed in the 50s and this is the model most librarians have in their minds, but the BIR model has huge drawback in that it is strictly a binary model. Either the document retrieved is relevant or it is not and hence under this model of Information retrieval the idea of relevancy ranking does not exist.
Learning point: Though we in the library world tend to use Boolean search and keyword search (lexcal search) interchangeably, it is important to realize that you can have keyword search or lexical search and not have strict Boolean!
For example, a TF-IDF algo (see later) may not necessarily follow Boolean search restrictions completely. Other times a search engine may be mostly following Boolean but if certain conditions are met (e.g. low number of results), the behavior might change (e.g. start stemming, expand synonyms)
For a real word example, my suspicion is that Google Scholar doesn't have a strictly Boolean search, but it is lexical, or keyword based because it's algo is still mostly based on comparing and matching keywords in query to documents.
Traditional classical keyword or lexical search tends to drop stop words and not take into account order of words using a bag of words approach.
This is opposed to semantic (occasionally called neural search) where matching is on the concept or meaning level and the current state of art techniques even considers order of words.
Ranking the results
Today it is unthinkable for academic searches today not to have relevancy ranking, this is where TF-IDF or Term frequency and inverse document frequency comes into play and provides different weights for matching each query term depending on TF and IDF.
Note: there are a few ways to calculate IDF, below shows one way based on log function.
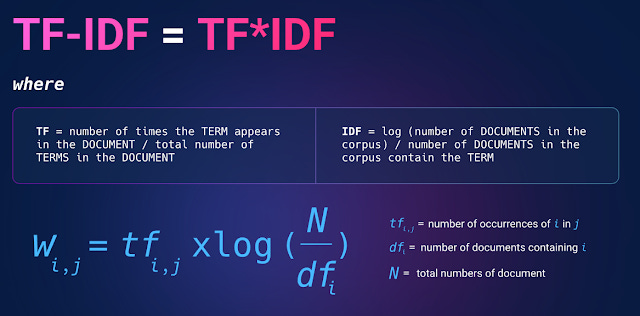
The idea is intuitive enough, if a query keyword that is matched appears in a document a lot (term frequency is high), the document gets higher ranked.
However not all terms are equally important. A common term that appears in a lot of documents (low inverse document frequency) would be less important to match than a specific term that rarely appears in documents (high inverse document frequency). This is represented by the inverse document frequency.
Extremely common words like "the", "is", "are" would of course have extremely low inverse document frequency since they appear in all documents. As such classical methods of information retrieval using "bag of word" type methods tend to filter them out as "Stop words". As you will see later more advanced techniques do not filter Stop words because they are able to consider order of words even for common words.
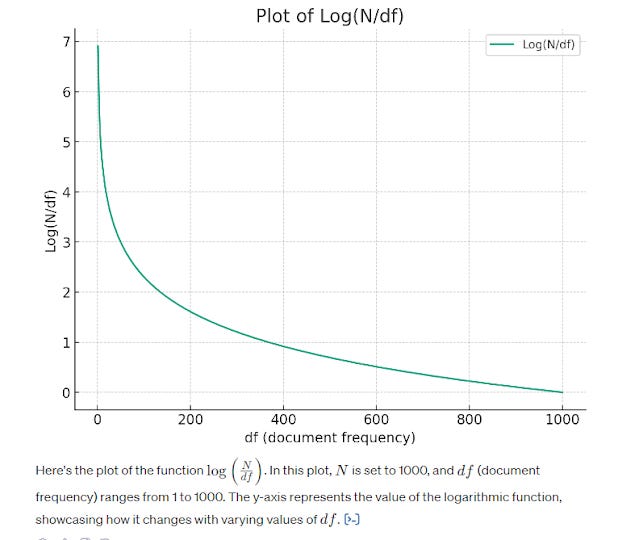
As the graph above shows, the more common a word can be found across all the documents (set to be 1000 in this case), the lower the score. At 1000, where every document has the word, the inverse doc frequency is at 0.
You can now get a TF-IDF score by multiplying both TF and IDF together,
You may be wondering that the tf-idf formula calculates one tf-idf score for each term. So Doc1 might have a tf-idf of 0.03 for <keyword1> , a tf-idf of 0.83 for <keyword2> and so on, but how do you combine them together? We will cover it in the next section on vector state models
BM25 is nowadays almost always used over TF-IDF, because it is a more refined version of TF-IDF as it takes into account length of document being matched, so longer documents in the set don't have an advantage.
However, TF-IDF/BM25 is technically a statistical model rather than a Boolean model so it can rank documents higher even if the document does not have all the query terms.
Today, many common search systems including academic ones based on elastic search combine the two methods - first doing a boolean match then ranking results based on TF-IDF/BM25 giving the best of both worlds, and this is what librarians expect to see.
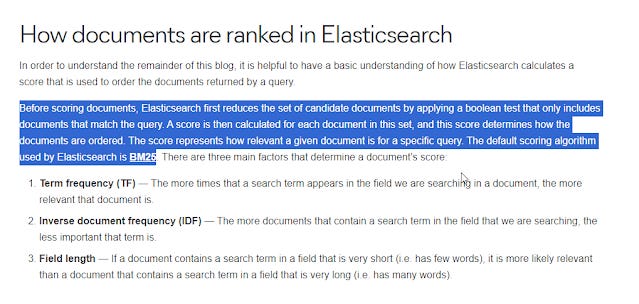
How documents are ranked in Elasticsearch
The predictability of Boolean/Keyword searches
I would guess most librarians when they say, "keyword search system" they operationalize it in their minds as a system that generates results that "make sense" or "can be explained". Such systems should be predictable enough to
a) allow a searcher to "explain" why a result was included or excluded based on search queries used
b) predict in advance when comparing two search strings, which should get equal or more results.
That said this isn't always 100% true because pure keyword systems rarely exist today, and academic searches may sometimes give "results that make no sense" because of additional "smart" features like search query expansion (typically rule-based) e.g., synonym matching that may trigger on different conditions.
There has always been a debate about how "helpful" search engines should be, e.g., PubMed ATM (auto term mapping) features and when they should be invoked and how to indicate it is happening
For example, years ago I remember a ruckus in a mailing list for a library discovery service where a librarian found that a search that was (A OR B) AND C resulted in fewer results than (A AND B AND C).
Eventually the answer given for this behavior was that the system had a rule where if the number of results fell below a certain threshold, it would automatically do stemming, and this can of course increase results.
There can be dozens if not hundreds of such additional rules that trigger depending on the situation that can occasionally make the search results less explainable.
Still at the end of the day, academic search systems even Google Scholar (but not Google as we shall see) are still mostly predictable and clearly lexical search systems based on matching keywords (though those keywords might be stemmed etc.).
Vector Space and rise of Embeddings (2010s)
Over the years, some search systems have tried other non-Boolean methods, like ones based on Vector Space models.
The idea here is you represent terms and documents as vectors in a multi-dimensional space and use a function like Cosine similarity to calculate the similarity between them.
One way to think about this is first imagine your universe consists of our two terms "dog" and "cat".
Each document can then be represented by two numbers X1, Y1, which represents how "dogish" the document is and how "catish" the document is.
For now, let's imagine X1 is just the number of times the word "dog" appears and Y1= number of times the word "cat" appear.
You can then plot both documents on a graph that is two dimensional as shown below.

You can see that Doc 2, is more "Catish" because it has a lot of "Cat" in the document and relatively little "Dog" and vice versa for Doc 1.
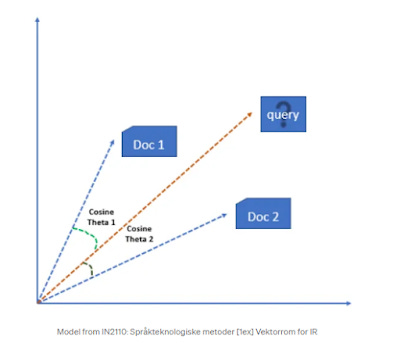
The search queries can of course be represented in the same way. A technical term is to call these representations a "vector".
The trick now is to ask for a given query above, which document (vector) is the closest or most similar to the query (vector). In the above image should the query retrieve Doc 1 or Doc 2?
A simple idea is to figure out whether Doc 1 or Doc 2 is more similar to the query is to look at the size of angles between them. By calculating the cosine between the angles of the query and each doc, one will get a score from 0 to 1. If the angle between the two vectors gets smaller and smaller, the cosine of the angle will approach one and if the angle becomes bigger and bigger it approaches zero. At 90 degrees, similarity drops to the lowest point or 0.
You may be wondering; this isn't a realistic example since documents have far more than two words. The beauty of this idea is you can extend this to more than just two terms or even three terms. If there are N terms you are representing, you can go into "N dimensional space". While human minds can't visualize more than three dimensions, math works the same!
Below shows the actual formula.

Below shows an example of documents represented with multiple dimensions (one for each word) using a document-term matrix.
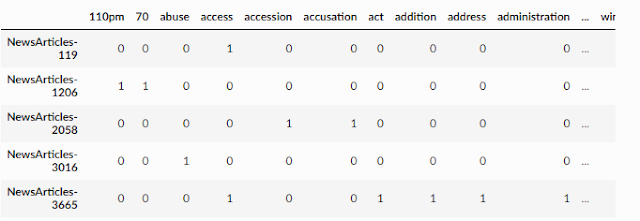
Document-term matrix
In the above matrix, every row represents a document, and each column represents a word. There are many ways to fill in the table, but in the above example, if a document such as newsarticle-119 has one or more words for the column it will be filled with "one", otherwise it will be "zero". But this seems a little crude, can we do better?
One could instead enter the number of times each word appears in the document (term frequency) but this does not consider how commonly the word is used across all documents (document frequency).
As such a popular way to improve on this method is to calculate a TF-IDF score and fill the cells with the TF-IDF score. This allows each cell to be not only a real number but also considers the weights provided by TF-IDF.
Technical note: Though BM25 can be seen as an evolution of TF-IDF, strictly speaking the BM25 is part of the probabilistic information retrieval models and not Vector Space models unlike TF-IDF. Probabilistic information retrieval models rank documents based on the probability that the document is relevant to the query.
BM25, specifically, is an extension of the Binary Independence Model (BIM), which is one of the earliest probabilistic models used in information retrieval. BIM operates on the assumption that the presence or absence of each term in a document is independent of the presence or absence of any other term in the document, given the relevance of the document. BM25 refines this approach by incorporating term frequency (how often a term appears in a document) and document length (the length of the document in terms of the number of words) into its relevance scoring formula. It also uses inverse document frequency (IDF) to account for the fact that some terms are more informative than others across the document collection.
BM25 can be considered within the broader context of the vector space model (VSM) framework, especially when we think about how documents are represented and compared to queries. However, the theoretical underpinning of BM25 itself is rooted in the probabilistic retrieval model and fundamentally differs from the traditional vector space model in its approach to document scoring and ranking. The ranking function of the probabilistic models is grounded in probability theory, while the ranking function of Vector state models — cosine similarity — is grounded in vector algebra
Clearly this method has disadvantages. Currently each document will have to be represented by a long series of numbers (one for each word in your collection!), this is clearly inefficient.
Is there a way to compress the same information in a shorter series of numbers? For example, you could imagine if your document has the words "cat", "feline" instead of having one column for each word or representing the document with two separate numbers, just use one instead since they mean roughly the same thing! If there is a automatic way to do this, the columns would represent meaning or semantics as opposed to just words.
Methods like Latent semantic indexing (LSI)/ Latent semantic analysis (LSA) in fact try to do this by trying to uncover hidden or "latent" meaning/groupings in your documents.
However, my understanding is that it was only after 2013 with the invention of techniques like Word2Vec/GLove etc that was the first sign that semantics or meaning could be captured and represented by what is now known as embeddings vectors which is also a series of numbers
The main difference is compared to the earlier vector space models used the words directly to represent as vectors, these newer models use embeddings or representations of words/tokens. How are these embeddings learnt? Though self-supervised machine learning of a lot of text as we shall see.
In today's context, words are not used directly but text are converted first into tokens, a process known as tokenization. There are various ways to do tokenization such as Word Tokenization, Subword Tokenization (currently state of art) and even character based Tokenization. Hugging face has good tutorials.
Like earlier Vector space models, you express both the query and documents as a vector embedding and you can calculate how similar embeddings are using various similarity measures such as cosine or dot product.
But what makes us think the embeddings capture meaning?
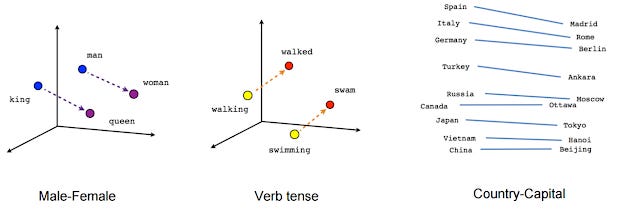
If you look at the positions of the embeddings, they seem to have some logic to them
Embeddings consist of thousands of numbers for each "word", actually tokens (one for each "word" in the "dictionary"), so they are technically represented in multi-dimension space, the diagram above has "squeezed them down" to just 2d.
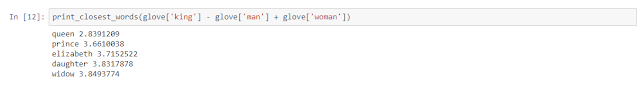
Mathematically you could do something like
vector(‘king’) - vector(‘man’) + vector(‘woman’) result is close to vector(‘queen’).
One major difference between embedding and the traditional vectors created using TF-IDF is that embeddings have a fixed length and do not depend on the number of unique words in the collection. This is because each number in the vector doesn't represent a literal word, but is a representation of some concept or meaning.
That said embedding vectors are almost impossible to interpret, since machine learning is used to automatically map latent or hidden meaning to each of these numbers in the vector (equal to columns in the the document-term matrix)
How are basic embeddings created?
Part of the issue with searching is the problems of synonyms, how do we know when you search for "cars" you are also searching for "automobiles"? Being able to look for words that are semantically "close" is clearly very useful.
So, you can see how an automated way to train embeddings or representations for words can be especially useful.
But how are such embeddings created? In general, the idea of embeddings is that similar words tend to be used in very similar contexts. Intuitively, if you have a sentence with a missing word and two words X or Y can be used in place of the missing words and this is true in other contexts, X and Y are somewhat related in meaning and should have very similar embeddings.
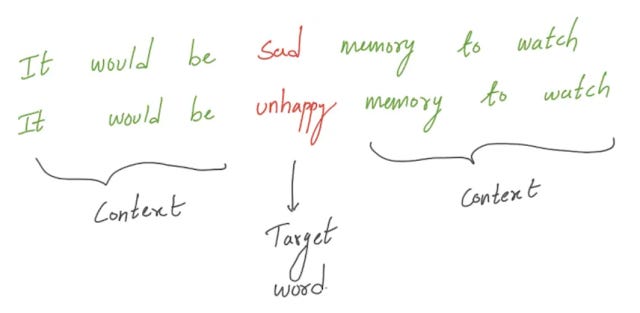
In the example above both "sad" and "unhappy" would fit into the sentence and hence should have similar embeddings and meaning.
This is the idea of Word2Vec embeddings, where a deep learning neutral net model is fed a large chunk of text and is used to learn how to predict such patterns.
Below shows part of the training data that is used to train using the Continuous Bag of Words Model.

It shows how you can use a chunk of text to train in a self-supervised way, where the aim is to predict the target word (in red above) from the context of the words in green. This particular training uses context window = 2, so it always tries to use the context of 2 words in front and behind the target word (if there are 1 or 0 words in front of target, it used that)
Given say the context
"number", "of" <target> "words", "to" , the neutral net should be trained to predict the target word "surrounding".
Once trained this embedding model will have weights that can be used to convert any text into embeddings.
Side-note on Sparse embeddings vs Dense embeddings
You will occasionally see TF-IDF/BM25 referred to as sparse embeddings and word2Vec and more advanced embeddings in use today as dense embeddings. This is now easy to understand.
Sparse vectors typically use the bag of words approach, where you either use raw count or using TF-IDF).
This of course results in a very long vector (high dimension space) with mostly zeroes in a Document-term matrix since you will need one column for every word (token) in the dictionary and most documents will not have a high % of the words, so for each row, most cells or elements will be zero, hence "sparse" embedding since most documents will be represented by long string of numbers mostly zero.
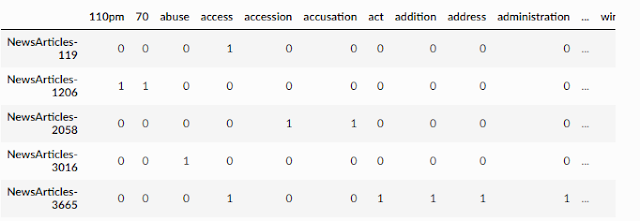
Sample example of a document-term matrix
Dense vectors on the other hand have shorter length vectors (lower dimension space) and much less of the vector elements are zero since each dimension represents a concept or meaning and most or more cells in the doc-term matrix will be non-zero.
This has already mentioned, is usually achieved nowadays by using machine learning to learn representations so similar words have similar embeddings.
That said older ways to create dense vectors did exist and used methods like Latent semantic indexing (LSI)/ Latent semantic analysis (LSA) but did not give much performance gains.
So, one way around this is to do a two-stage search and reranking system where the sparse retriever first retrieves a large set of results, and this is reranked using Dense reranker. This ensures results/documents retrieved are explainable even though the order of them might be a Blackbox.
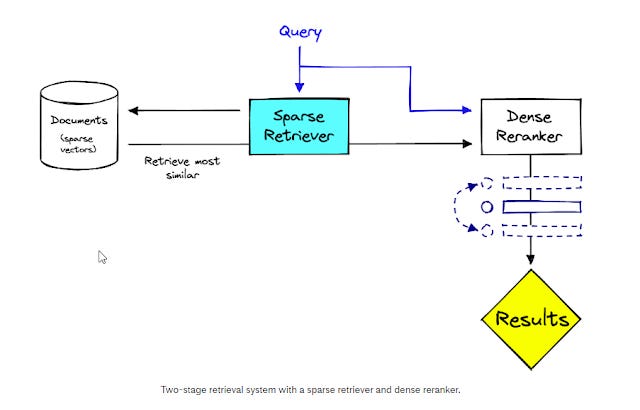
Two stage retriever using sparse retriever first than dense reranker
The most significant advantage of SPLADE is not necessarily that it can do term expansion but instead that it can learn term expansions. Traditional methods required rule-based term expansion which is time-consuming and fundamentally limited. Whereas SPLADE can use the best language models to learn term expansions and even tweak them based on the sentence context. See here
Rise of contextual embeddings from transformer based large language models (eg BERT), 2018-
The main issue with older methods was that it couldn't capture meaning (semantic) automatically, and Word2vec and similar embeddings at the time helped. But it still didn't fully solve the issue of taking into account word order in documents or queries.
Take the query (this example was taken from Google's announcement of they use of BERT in 2019)
2019 brazil traveler to use need visa
the systems needs to consider the order of the query and understand it is about someone from Brazil going to US and not vice versa. To solve such issues, every word counts, and you the latest embeddings (from 2018 onwards) far improve on Word2Vec type embeddings due to improvements like the self-attention mechanisms and positional embeddings.
In fact, Word2vec type embeddings were just the beginning. Transformer based large language models (invented in 2017) were found to produce even better embeddings due to improvements like the self-attention mechanisms and positional embeddings allowing such systems to take into account position/sequences of words.
The most famous example of these types of new embeddings was introduced in 2018 was named Bidirectional Encoder Representations from Transformers (BERT) and was quickly adopted everywhere leading to huge gains in state of art.
Another advantage of embeddings is because they supposedly capture semantics you can create language agnostic embeddings that work across different languages! (See also)
Further improvements on contextual embeddings
But even BERT models are just the beginning. Today besides BERT family of models, there are many other advanced embedding models that have been further fine-tuned to do well for semantic search and reranking including
Variants based on BERT / SBERT - MiniLM, mpnet-base family (light weight high performance)
Variants using Google's T5 as a base
GPT based e.g. OpenAI's text-embedding-ada family, Cohere embedding etc
Domain specific trained on scholarly publication and pretrained with citation signals - e.g. SPECTER by Allen institute for AI and specific BERT models finetuned with papers e.g scibert, finbert or even vendor specific ones like Dimensions General Science-BERT
Here's a brief over-view on further improvements
Firstly, how good embeddings are (even ones from state of art Large Language Models) highly depends on the data that was using to train on. For example, if journal articles were used mostly for training the embedding models, it may not work well when used to find similar articles in the domain of newspapers!
In fact, most standard BERT base models were/are trained at least partly on Wikipedia text, so it may be possible to create or fine-tune embeddings models more suitable for specific domains like academic articles or even specific domains like Chemistry (ChemBERT), Finance (e.g. FinBert), SciBERT etc. SPECTER by Allen institute for AI even considers citations!
In-domain training to further push the limits
BERT models have a pretraining phrase where they are trained using the masked language model where it [mask] words in text (somewhat like the CBOW from Word2vec but not exactly the same) and Next Sentence Prediction Tasks.
Unlike the GPT models, BERT models are encoder only models, and they do not generate anything (because they lack a decoder) but embeddings. You also are expected to generally do further fine tuning on the model for the task you want to do with the BERT model.
In this case, how or what should you fine-tune?
Arguably embeddings/similarity alone based on masked language tasks are not always sufficient to get relevant results just from pretraining. Also, even if query passages and document passages are similar it does not mean they are the answers or documents needed. Could you not improve performance further by training the model further on specifically labelled documents and queries?
This is where supervised learning techniques on large datasets with queries and answers like MS MARCO are done to push the limits of performance further.
However, this is highly domain specific and not all domains have the needed labelled data to do supervised learning and there is now active research in finding self-supervised techniques like Negative sampling/contrastive or even using of Language models to generate synthetic data for such domains.
It is also unclear as you whether fine-tuning on domain specific examples might hurt performance if it is used out of domain.
These days you are not even limited to using embeddings from Encoder only models like BERT but even popular GPT type Decoder only models like OpenAI and Cohere provide embeddings. I suspect for quick and easy use OpenAI's text-embedding-ada-002 is extremely popular! These embeddings are generated from even larger text sources than BERT models but it's unclear how they match up against BERT domain specific models.
Bi-encoder vs cross-encoder
Another important concept to understand is the difference between a bi-encoder and a cross-encoder. This impacts how the query, and the documents interact.
The example shows an example of a system trying to decide if two sentences are similar using a sentence embedding based on BERT. In the context of search one sentence would be the query, one sentence would be from the potential matching document.
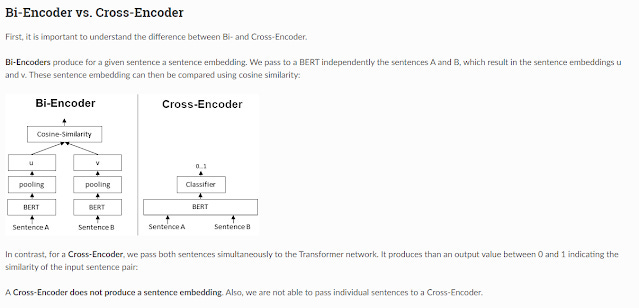
The most common type of system implements a Bi-encoder where both sentence A (the query) and sentence B (potential matching sentence in document) will be converted into embeddings separately before they are checked for similarity using a function like Cosine-similarity/dot product.
The main advantage of this method is that you can precompute the embeddings for all the documents of your database in advance and store it in a vector database. So, when the search is run, you just need to convert the query into an embedding and then you can use methods that are reasonably fast (approximate nearest Neighbour) to quickly find the embeddings that are closest.
Now consider the alternative cross-encoder method. The Cross-encoder has an even better performance, since you push both query and document into one model which allows the model to take into account interactions between both items (as opposed to the bi-encoder method where you create two separate embeddings and allow interaction only via something like dot product).
But cross encoders are clearly more difficult to do computationally since you cannot precompute the embeddings for documents like for a bi-encoder. If the database has 100k documents, you need to do all 100k documents embeddings + query pairs on the fly during the search!
A workaround to get the best of both worlds is to use some other method to first cut down the number of top candidates results then do a rerank of these top candidates using a cross encoder.
One obvious method is to use the biencoder to grab the top 100 then the crossencoder to rerank.
There is also a late interaction model eg COLBERT (Contextualized late interaction over BERT) that is in between the bi-encoder and cross encoder.
Is the newer Dense embedding better than Sparse embeddings?
In general, dense embeddings are better than standard sparse embeddings in specific domains but tend to be weaker if used out of domain according to the 2021 BEIR paper.
So for example, to take a crude example if your dense embedding is trained (either supervised with labelled pairs or self-supervised) on journal articles and you are trying to do newspaper searches it can fail.
Indeed a 2021 paper found that accross a wide variety of settings(domains and tasks) such as in the BEIR test, BM25 is a very strong base line to beat. In short, if you want something to work well across a large variety of different domains or are unable to be sure the old school BM25 is your best bet!
However, this may be a less relevant problem in academic search where the domain tends to be quite restricted.
Besides technology marches on and a June 2023 blog surveying the results from the same benchmark concludes that in 2021, while BM25 was a very strong baseline it is no longer a clear winner in 2023 and is clearly beaten by the latest dense embedding like E5 and newer sparse embedding methods like ColBERT (Vespa’s ColBERT), SPLADEv2, ELSER
Dense vectors are effective for encoding local syntactic and semantic cues leveraging recent advances in contextualized text encoding (Devlin et al., 2019), while sparse vectors are superior at encoding precise lexical information.
Hybrid approaches also help reduce the weakness of Dense embedding approaches of being weaker in out of domain tasks (which BM25 excel at).
There are multiple ways to do so from
Dense first search — Retrieve candidates using a dense model and rerank using the sparse model
Sparse first search — Retrieve candidates using the sparse model and rerank using a dense model
Hybrid search — Retrieve candidates from both lists and combine them as a post-processing step
It seems the current evidence (2023) and industry practice favours a hybrid approach as doing either Dense first or Sparse first risk missing relevant documents, see this article for discussion of why
Another more practical issue is that users often want control and predictability in their searches, what happens if they search and also require that the documents be after 2020 only? Or if they want an exact match in the title field only?
All this points to the need of a hybrid system using both semantic and lexical features…, See also
Why I think semantic search might be finally coming for academic search
It is interesting that when you look at academic search, even Google Scholar, their search results are still mostly traditional keyword based. Surely, the search isn't 100% explainable sometimes, but there is still an expectation that most of the time, you will not be wondering why a certain document appeared in response to your search (with some fogginess due to stemming, synonyms etc.) and documents are still matched one on one with query keywords.
I believe this might be changing for the following reasons.
Firstly, the popularity of ChatGPT means people are getting used to searching with natural language further reinforcing the trend Google began. While I don't believe keyword searching will go away that quickly, I believe there should be an uptick in natural language searching and if that happens the most natural way of supporting it would be via semantic search.
Secondly and most importantly, I think Semantic Search has reached a point where they match and often outdo the performance of traditional keyword-based/Boolean search.
I've often given an example of how in some search queries - Elicit.com trounces Google Scholar in finding relevant papers even though Google Scholar has a huge advantage in terms of the size of the index, particularly in terms of full-text.
Take the example below - how to find seminal works
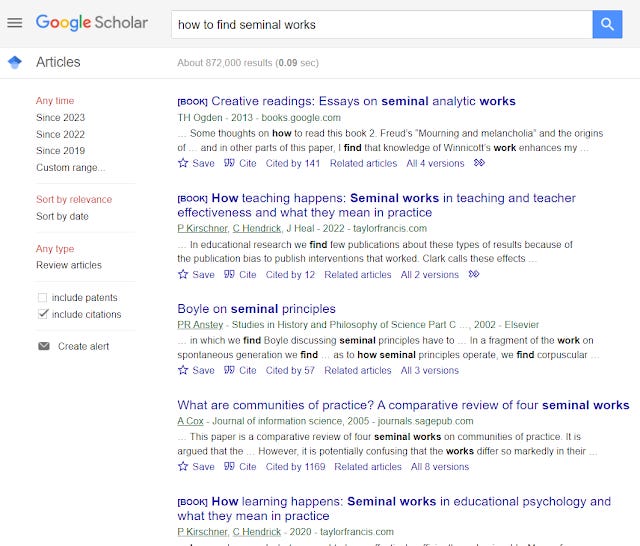
Google Scholar totally fails here and the words that are highlighted in the snippets tell you why. It clearly doesn't get the intent.
Elicit.com does way better (but not perfect).
Defenders of Google Scholar might say you could get better results if you used quotes around "seminal works" (only slightly) or that you use just drop "how to" (still bad). In fact, I found that by searching with query identify "seminal works" the results get better, but it does show how sensitive the search is to the right keyword!
By comparison Elicit.com's results do get slightly better if you try searching identify seminal works, but in general the results are less sensitive to minor changes like quotes, identify vs find which makes sense given it is trying to match semantics.
JSTOR generative AI pilot now provides a experimental new mode doing Semantic Search
For reasons already mentioned, academic search engines are often black boxes, and it is hard to tell if they are using Semantic Searches.
JSTOR generative AI pilot however makes it easy. They recently launched a new "experimental search" that "understands your query and provides more relevant results, even if you don't use the exact words."
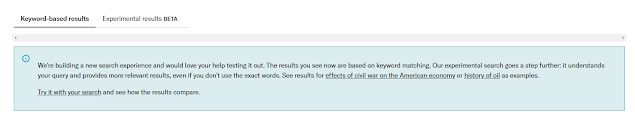
"They use a variety of factors to understand the meaning of your query and the relationships between different concepts. It helps you find what you are looking for, even if you don't use the exact words."
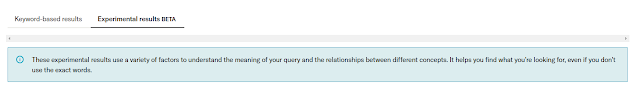
Clearly some sort of semantic type search being used. I haven't given it a full workout but so far, I am quite impressed.
One of the issues using such semantic search tools is that it is unclear to me how we should search.

If it is a straight-forward strict boolean and lexical type search clearly only searching conventional keyword style is the way to do and trying to type in natural language is going to fail.
But given many of the new academic search can now give direct answers and with the influence of ChatGPT, it may start to get a bit confusing about whether one should type in natural language or even do long prompt engineering style inputs!
In general, if there is truly a semantic search implemented it should be able to handle single natural language inputs and even conventional lexical search might if the stop words are dropped. Prompt engineering style inputs I think are less likely to work particularly for straight out search engines that can't do any other task but search.
In the case of JSTOR generative AI, I tried in both natural language and keyword style.
JSTOR generative AI - Is there an open access citation advantage? (natural language style)
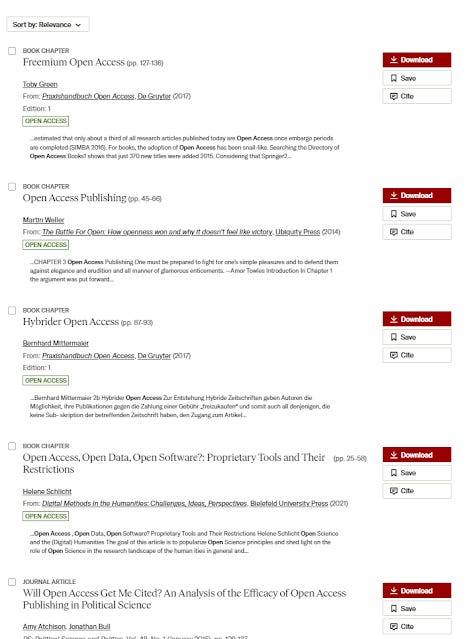
First, we look at the traditional status quo search, the first four results are bad.

The results from the experimental, Semantic Search are much better with the first two results being clearly relevant.
But perhaps, the natural language search is causing issues. Let's try keyword search query
Open Access citation advantage

The results from the default search is just as bad.

Interestingly, the new experimental search result is still superior to the default search but seems to be worse than if we did natural language!
In case you are wondering, I actually tried a few examples and almost every time the new experimental search gave better results or at worse similar results! It's really interesting....
Implications of Semantic Search feature and JSTOR's clever new feature
I know a lot of librarians, particularly evidence synthesis librarians reading this are probably very concerned. Already they are worrying over the lack of precision due to auto-indexing of MeSH...
In fact, I have had librarians come up to me after talks asking me advice for how to advocate to database vendors to keep lexical search (or more accurately power user boolean search functionality). and not replace them with semantic search.
Evidence synthesis librarians clearly desire the precision and predictability that lexical boolean search provides.
That said JSTOR generative Ai pilot has implemented a pretty clever feature to help mitigate one of the drawbacks of using semantic search over keyword/lexical search.
With pure lexical search, you can just look at the search engine context and look at the highlighted words to quickly determine if a paper is relevant. But this works less well with semantic search since it may not always match the keyword in your query term.
As a trivial example imagine you searched "automobile US", the semantic search recognizes the "US" to mean America here and gives you documents due to that. But straight forward keyword highlighting won't work here. This is just a toy example, but you get the idea.
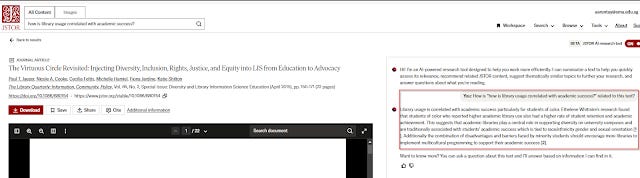
JSTOR generative AI tries to mitigate this issue in a clever way I have not seen anyone else do.
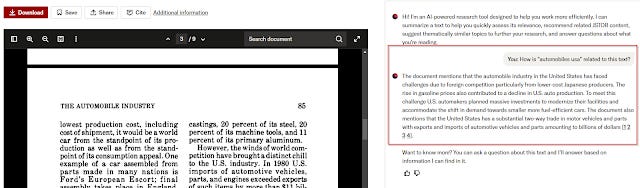
Like many other tools, JSTOR implements what I call a "Talk with PDF/content feature" where you can ask predefined questions about the full-text in one document including as of time of writing
What is this text about?
Recommend topics?
Show me related content
Ask a question about this text <open ended for you to type>
This isn't too novel. What is interesting is that when you click in from any search result page in JSTOR it will automatically prompt the language model
"how is <query> related to this text"
This is very clever and assuming it is reliable you can quickly tell if a paper is relevant or not!
I have tried it quite a bit so far it seems quite accurate. I love the fact that despite the positive phrasing of the prompt "How does <query> relate to" implying we expect the answer to be yes , it is often ready to say "No it is not related"!
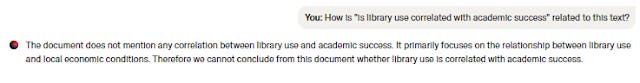

It occurs to me that in the usual case where it says a document is related to the search query, it provides evidence where you can mouse-over and see the text for verification purposes.
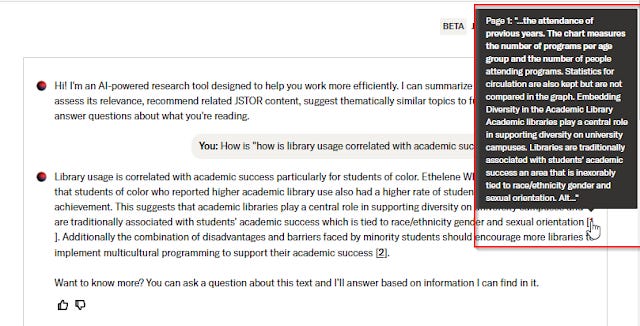
However, when it says it is not relevant, there is no easy way to check to see if it's not a false negative!
Conclusion
There are two general ways to find papers. First is good old fashioned keyword search. Second is using citations. For the second method, I have tracked the rise of tools like Research Rabbit, Connected Papers, Litmaps from the 2020s that make it much easier to use this method than before, the fruits of a successful campaign to make scholarly metadata openly available in machine readable format (particularly citations/references).
Now we see the rise of a third way, semantic similarity and in fact tools like litmap are adding some semantic function based on similarity though currently only on title and abstract.
Hopefully support can also come from the "semantic side", so that one can prompt or do searches like. Take paper A, find the 5 most semantic similar papers, extract all citations to these 5 new papers and dedupe or vice versa.
Another interesting thought occurs to me. Tools like Elicit, scispace are now charging per use/credit. We obviously understand why since currently the inference cost of running a search the cost isn't trivial whether it is paying for API calls or running the compute on your local or remote cluster.
This brings to mind the old days of Dialog where librarians do mediated searching for users. In those days, where you build up search strategies with nested boolean of the form
(A OR B OR C) AND (D OR E OR F)
where A,B,C and D,E,F are synonyms are must haves because of the need for efficient searches (since you are charged by usage). Yet the irony is many of these tools support Semantic Search so you can't even do nested boolean!

