Using oaDOI & Crossref event data API to calculate your institution's open access citation advantage
As a generalist with hands in many pies, I'm prone to throw around terms I barely understand.
API or Application Programming Interface might be one of them. But learning about APIs are important as we are using it whether we know it or not. Recently, I have started to get comfortable with them , at least in terms of pulling out the data and parsing the JSON output.
In this blog post, I will show you how to use 2 APIs, the oaDOI API and the new CrossRef Event Data API to calculate the citation advantage of open access articles for works by authors in your institution.
But even if you don't care about that, the real intent is to walk through with you how to use these 2 APIs even if you have never touched an API before. With this knowledge you should be able to handle the basics of pulling out data using APIs.\
This is also an important skill for doing research......
What is the oaDOI and CrossRef Event Data API?
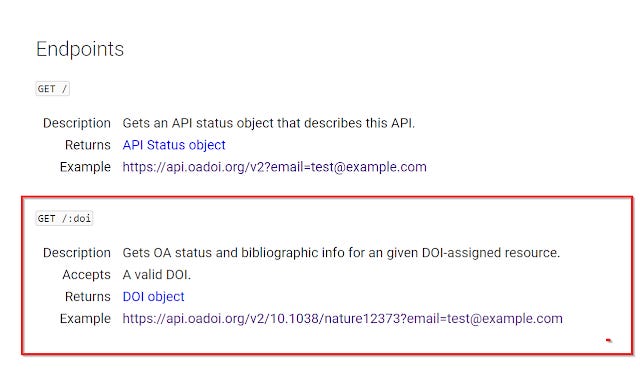
The oaDOI API is basically the service behind Unpaywall that allows you to automatically check for free articles. With the API you can query a Digital Object Identifier System (doi) and it will be able to tell you if a free to read version is available and where it can be found. It covers both publisher sites (gold and hybrid) as well as repositories both subject and institution (but see some limitations later).
The Open Access button API has similar functionality, the main difference if any is that it can take not just dois but also "PMIDs, PMC IDs, titles, and citations" but otherwise the method to handle it is similar to oaDOI API.
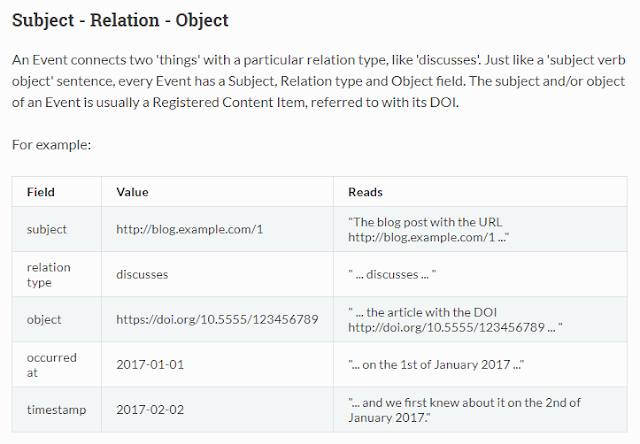
Another very useful new API is the CrossRef Event Data API. It is a very powerful API that helps you with altmetrics. The main use is that it allows you to tell if a certain doi has been mentioned or cited in Tweet, Wikipedia, Reddit, Blogs and more.
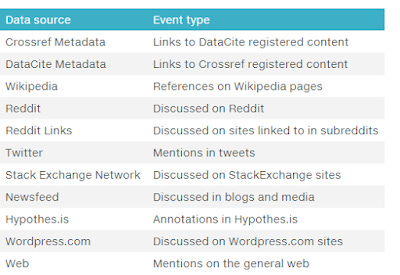
Some sources covered by new Crossref event data API
But again if you can handle oaDOI API using the method shown below, you can handle this new API.
The idea here is simple, using the data from the CrossRef Event Data API, you can study whether been mentioned or discussed in these sources is correlated with having a free copy/ or is well cited in traditional ways.
What I like about these 3 APIs already mentioned is that they are free and currently don't have any limits and you can freely use them without registering for API keys (oaDOI recommends use up to 100k per day). This might change of course in the future if people abuse the API.
Note : A lot of this is mostly inspired by the tutorial here but I have updated the instructions below to handle v2 of the oaDOI API and other additional ideas.
1. Get a set of bibliometric data with dois and suitable citation data
To begin, you need a set of dois. One easy way is to get them from a source like Scopus. In my example, I pulled out all papers written by authors from my institution for a range of years. It goes without saying you can use any other methods as long as you get a set of dois you want to work with.
If you are looking to do a study on the citation advantage of open access besides dois, you will need to get citation metrics as well. For my example, I am using Scival to get Field-Weighted Citation Impact (FWCI) to enable comparisons across different disciplines and years.
So I now have a spreadsheet with columns for dois, various citation details such as title, year of publication, journal title and field weighted citation impact (FWCI)
Now for the tricky part, for each record , I need to find out if there is a free version for each item. This is where the oaDOI API comes in.
2. Working with oaDOI API using openrefine
If you are comfortable with R, the best way to work with oaDOI API is to use the roadoi R Library. But the instructions below will help you do the same without using R, instead we will use OpenRefine.
First, install and run the free OpenRefine. When Openrefine runs it will open a browser window with the web interface.
Once it’s loaded, click on
Create project , Get data from this computer , Choose file and browse and select the file of dois you have .
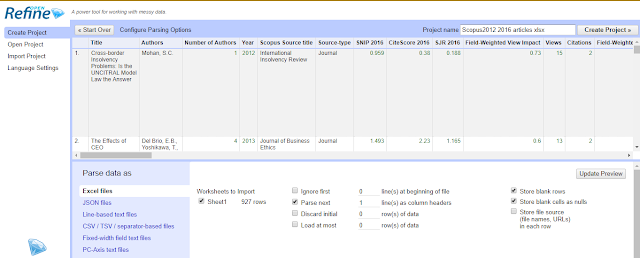
In most instances, openrefine will automatically recognise the file as csv or excel and the settings should be obvious.
Once you have checked everything is loaded okay, click on “create project”
Next, I usually like to check how many of these records I imported have dois. This is important as oaDOI API only works on dois. How many of your records have no dois or blank values in the doi field?
To do that, go to the
DOI column (click on the down arrow) , Facet , Customized facets , Facet by blank.
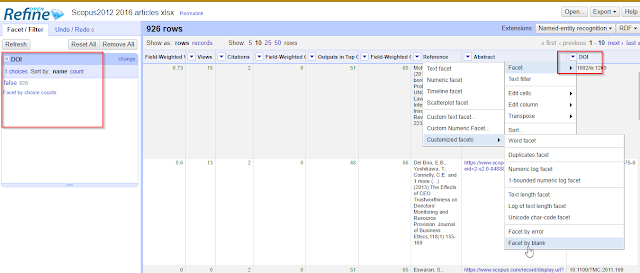
Facet the doi field by blank to check
I see from the side filter panel on the left, 926 records have dois(blank=false) and there are no records with do dois (blank=True). If you happen to have records that have no dois , just click on the value false to filter to only records with dois.
The next step is where the magic happens. On the DOI column (click on the down arrow), click
Edit column , Add column by fetching URLs.
In the expression space, type
"https://api.oadoi.org/v2/"+value+"?email=xxxx@yyy"
(replace xxxx and yyy with your email of course)
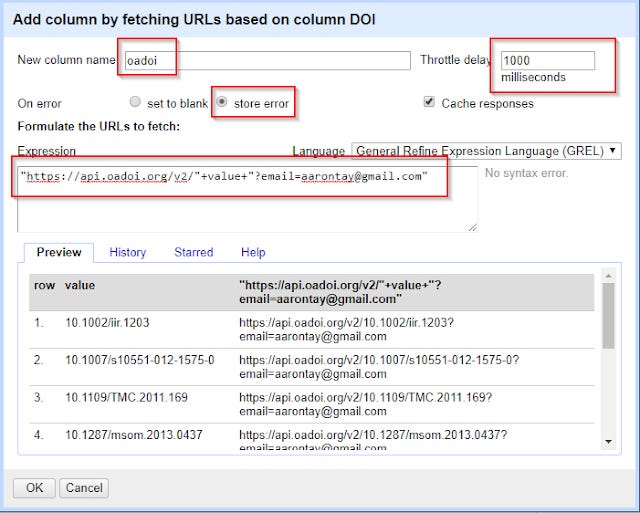
This constructs for each record a API call to oaDOI using dois. Don’t forget to name the new column, in my case I used oaDOI.
Tip : As I write this , there is a slight bug with the latest version of OpenRefine 2.7 where you may encounter a problem where OpeRefine will stall at records with errors (e.g. if you have non-dois). To avoid this problem, you should select "On error" "store error" rather than the default "set to blank".
What did we just do?
Basically we told Openrefine for each record please generate a different request to the API. Openrefine will replace "value" with whatever the doi is for each record.
So if for example, if the doi is 10.1002/iir.1203 for the first record, OpenRefine will send
http://api.oadoi.org/v2/10.1002/iir.1203?email=aarontay@gmail.com to the API .
Which will return a output that will be stored in the new column you just created.
The same will occur for the next record except the value portion (the portion in red above) will be another doi.
Most APIs require you to enter a API key to prove you can access it, but currently the oaDOI API just requires you to enter your email. No registration required.
How did I know this is the right string to send to the API?
Just read the documentation
How long will it takes for the processing to finish?
To speed up the process, you may want to adjust the "Throttle delay" down from the default 5000 miliseconds. This setting basically tells openrefine to time how fast requests are made to the API. The smaller the throttle the fast the work can be done, but many APIs have limits to how fast you can query them. For oaDOI, I've found 1000 miliseconds (or 1 second) to be a reasonable throttle delay.
3. Parsing the Json output - extracting "is_oa" key value.
If you set the threshold delay to be 1000 milisecond it will take appropximately 1 second per record, so 1000 records may take roughly 16 minutes.
Here's where it gets tricky. Most APIs return output in either JSON and/or XML. The question is how do you interpret them?
Okay I know the output looks scary particularly when all squeezed into openrefine but don't panic. What I like to do is to copy and paste the JSON to a JSON formatter like this one to make it more human readable.
Here's part of what you might see.

First let's ignore the whole block of text in "best_oa_location" and look at the rest.
The easiest thing to do if all you want to know if there is a free version is to try to extract the value for the field "is_oa". If the value for "is_oa" is true, there is a Open access copy found by the API. If the value is "false" oaDOI couldn't find anything.
So how do you extract the value of that field in Openrefine?
On the OaDOI column, click on the down arrow , Edit Column , Add column based on this column.
In the expression column try
value.parseJson().is_oa
Again remember to enter a name for the new column.
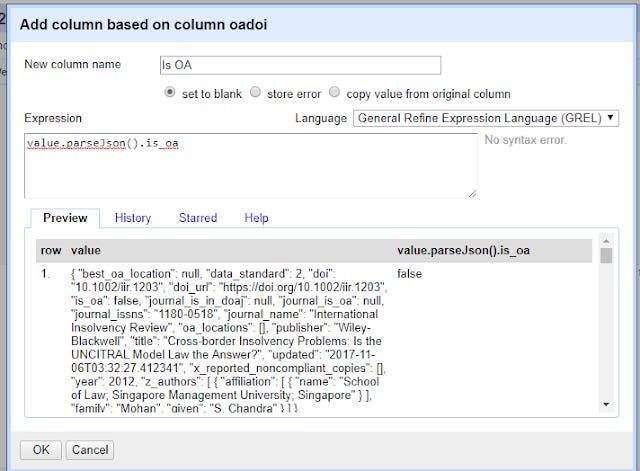
This looks simple enough right? You are telling openrefine, for the value in the cell, run the function parseJson() and grab the value from is_oa.
Slightly more advanced Parsing the Json output - extracting best oa location
But what if you want to grab more information? Like where the free copy is, and what version it is?
If you read the documentation, you know that the API will try to find every copy of the paper it can find. So one doi might have copies at a publisher site and a subject repository and a institutional repository and oaDOI will give you the details of all of them.
So the API will decide on the best version (usually the one at the publisher), and this will be in a block of results called "best_oa_location"
If we want to extract details related to that, what do we do? For example, how do we get the URL of that "best_oa_location" copy?
On the oaodoi column, do the same as before but in the expression column try
value.parseJson().best_oa_location.url
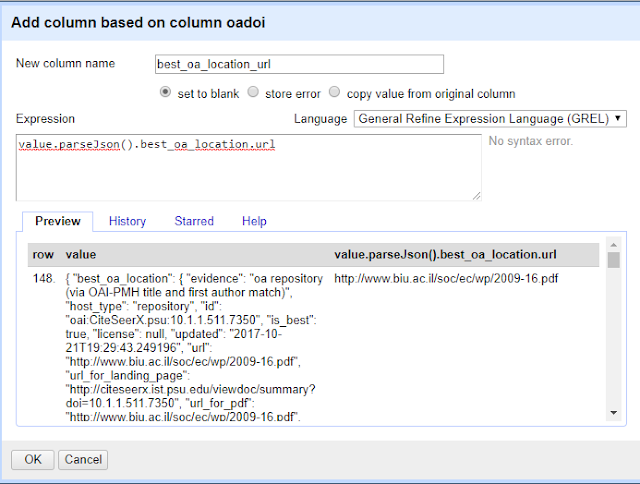
or if you want to see what version is available at the "best location" try
value.parseJson().best_oa_location.version
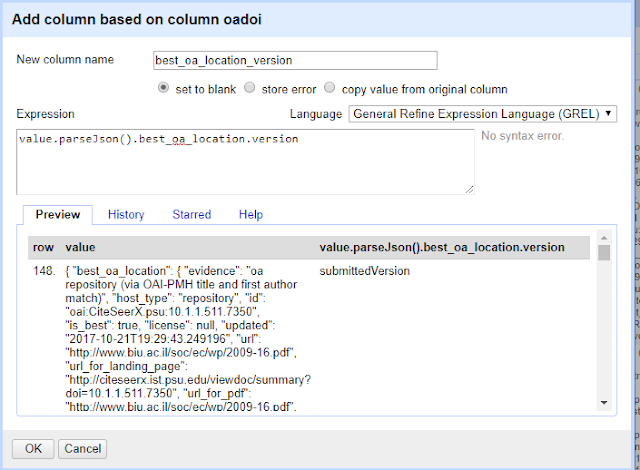
Note that in the preview window, you might see errors like the following.
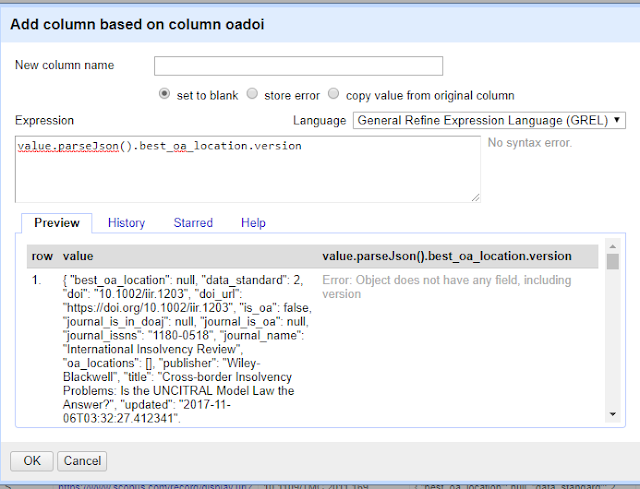
This is perfectly normal for records that have no free full text found.
Hopefully you are starting to see how this works. Can you extract other values from best_oa_location such as "host_type"?
Incidentally unlike v1 of the API, v2 of the api does not define Green OA or Gold OA. By using the values in "host_type" which can have values of "Publisher" or "Repository" together with other values like "journal_is_oa" you can make your own definitions.
Parsing the Json output - extracting other oa_locations
What about the other locations of other copies found?
This gets a bit tricky. First, notice that for each doi, the number of free copies that can be found varies. It can vary from 0 to a large number and each copy found generates more key value pairs.
So you need to specify which particular results you want. But where are those results?
The key here is to look at "oa_locations" notice it is followed by : and everything is enclosed in a square bracket [ ].
To make it easier to see, I've collapsed everything else , so you can see it.
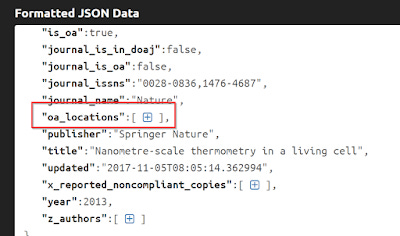
All the information about other copies are found in what is called an "Array", which you can identify because it is nested in a set of square brackets [ ] . If you expand this further, you can see there are 2 other blocks representing the 2 other locations.
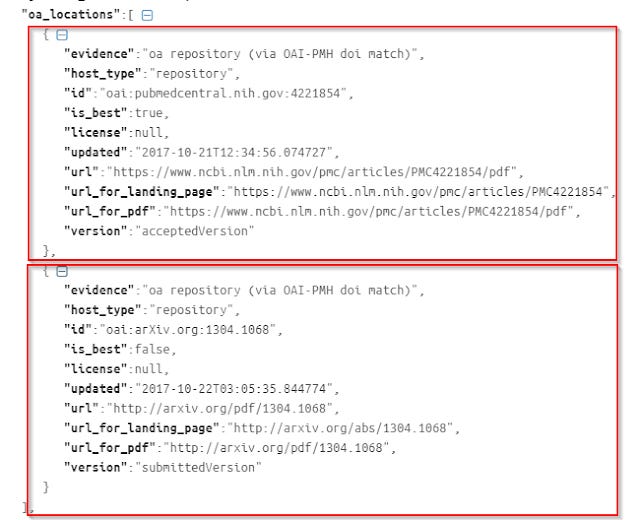
So unlike say "is_oa", where there is only one value, "oa_location" has multiple (in this case two) results.
In this example, you can see there's one copy in Pubmed Central (pmc), and another in arXiv. So how do you extract say the URL for the first or for the second? You can't just do
value.parseJson().oa_locations.url
because Openrefine doesn't know which url are you referring to. There are two of those in "oa_locations".
So instead, using the same method as above the expression you should instead do
value.parseJson().oa_locations[0].url - for the URL of Pubmed Central version
and
value.parseJson().oa_locations[1].url - for the URL of arXiv version
Ordering of results
What's the main difference? Whenever you run into an array, aka when you see blocks enclosed by square brackets, it means you know there will be multiple results and you need to tell Openrefine which result you want.
So you add a [] which tells you which result you want.
[0] - tells the system you want the first result, while [1] - gets you the second result.
Yes the numbering starts at 0 for this, don't ask me why.
4. Do a t-test of the average field weighted citation impact of the articles with open access versions and those without
Once you have parsed out the JSON output to allow you to tell if they have free copies or not the obvious thing to do is to split your dois into 2 groups those with free copies and those without.
Find the average field weighted citation impact of these 2 groups and run a t-test using whatever method you prefer, either with Excel, SPSS, R or even a simple t-test form calculator.
Hopefully you have found a signficant and meaningful citation advantage
5. Improving your results by enhancing with Institutional repository results
You may be wondering how accurate oaDOI is at detecting free full text.
Based on a recent study it generally has a high precision but relatively low recall. In other words, if it says there is a free version, it is usually correct (roughly 96.6% of the time) but it often misses free copies (it only finds 77% of free articles).
In particular, my experiments have shown that a lot of the missed copies can often be from subject repositories. This is because while it's relatively easy to cover and detect free copies in publisher sites and centralised big subject repositories like Pubmed central, there are thousands of institutional repositories with differing standards of indicating full text.
My understanding is oaDOI and other similar services rely heavily on BASE (and for some CORE) aggregators to detect copies in institutional repositories. The problem with this is not that BASE hasn't harvested your institutional repository (it's likely it has) but it may not be able to determine if the data there has full text or not.
The problem varies from institution to institution, but here in Singapore, while most records in the institutional repositories are in BASE, BASE is only to detect less than 4% of the full text as full text! Another issue is that your institutional repository might not have a doi attached.
Even without this weakness, oaDOI is likely to get less free full text than Google or Google Scholar. This is because it ignores (on purpose) free full text in ResearchGate and Academia.edu as well in non-publisher or repository sites. So it will miss out free full text posted on University personal home pages.
You may not have this problem, but I think it's prudent to try to improve the reliability of your results by supplementing the results you get from the API by checking on the ones that oaDOI is unable to find free copies vs your institutional repository's list of full text.
You might also consider using the Open Acess Button (OAB)API as a double check and by now you should be able to do this in almost the same manner, but it's can't clear to me how reliable OAB is.
Latest : Just as I was going to relase this blog post, oaDOI has announced they are no longer relying on BASE for detecting insitutional repository copies. They are now using their own harvester which avoids the difficulty of detecting free full text in institutional repositories.
"There are a few challenges to using BASE data. One of the most important, we already solved: they frequently do not know whether a record actually has fulltext available. So we go to the IR and actually check. That way, when we say there’s an OA copy somewhere, we can be sure there really is. We have downloaded the PDF and we know for sure." (oaDOI mailing list, 4th Nov 2017)"
Still it will take some time for this to take effect, so I recommend doing a double check.
Working with the new CrossRef event data API
CrossRef is a pretty well known organization in scholarly circles due to the ubquity of dois, though some people even librarians mistakenly believe CrossRef is in charge of all Dois, when in truth they are several DOI Registration Agencies (another you might have heard of is Datacite) and they are just the biggest and well known one in Scholarly circles.
They have a pretty well known CrossRef API, which has many uses, chief of which is to get metadata contributed by their members (mostly publishers).
But the API I'm writing about is a different one, the CrossRef event data API which is currently in beta.
This is a much more complicated and full featured API compared to oaDOI API, but backedup with a huge amount of helpful documentation.
So how does it work?
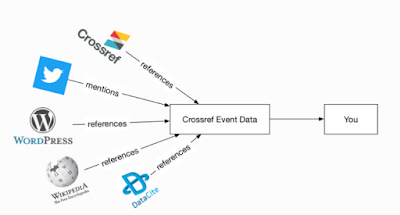
https://www.eventdata.crossref.org/guide/introduction/
I spent a couple of hours reading the documentation and by the end of it my brain was swimming. It is very very detailed.
For instance besides checking if a doi has been mentioned on Twitter, you can query the "evidence log" which allows you to tell when exactly the event was supposed to happen and when the data for that event was collected.
Still I got the basics enough for a first try, this is my current experimental attempt.
Constructing the web query for API
As before, On the DOI column (click on the down arrow), click
Edit column , Add column by fetching URLs.
In the expression space, type
"http://query.eventdata.crossref.org/events?rows=10&filter=obj-id:"+value
https://www.eventdata.crossref.org/guide/introduction/
I'm basically saying to the API "show me all events where the object is the doi which is the value in OpenRefine."
Parsing Json

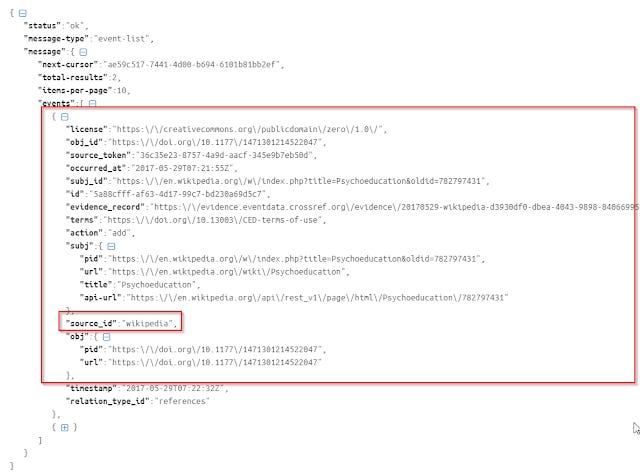
Parsing the Json output is a bit more tricky than earlier examples. Like the case of "oa_locations" for oaDOI where one doi might have many possible copies, in the CrossRef data API, an object DOI, might have more than one event associated with it.
As seen above, this example has 2 events associated all nested in an array. You can tell because it's in square brackets [ ] . Each event will both have the source_id field (which tells you if it is from Wikipedia, Twitter, Reddit, websites etc)
"Source_id" of first event.
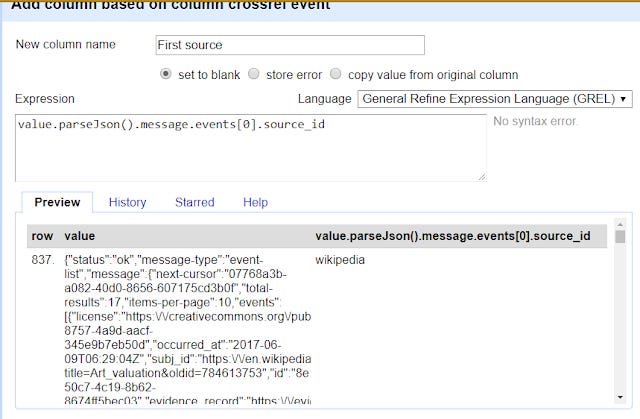
Using the earlier example, you know to extract the first result for "sourceid" (which tells you if it is from Wikipedia, Twitter, Reddit, websites etc) you simply do
value.parseJson().message.events[0].source_id
If you wanted the second result's source just do
value.parseJson().message.events[1].source_id
Other interesting fields to extract are "occurred_at", "pid", "subj:url".
Parsing Json with foreach
But what happens if you want to extract all the fields for each result into one cell? How do you do it?
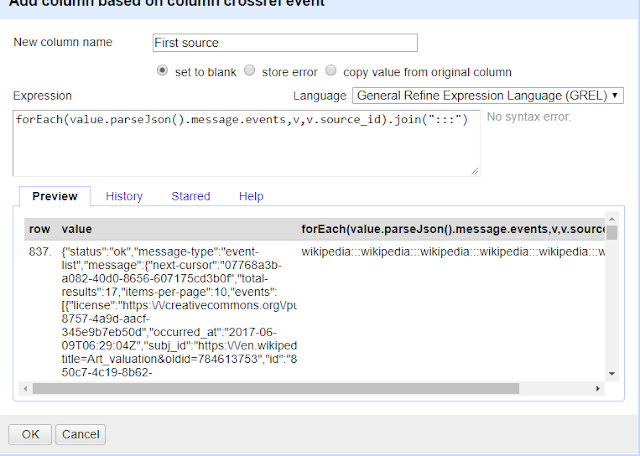
For example, you want may want to extract all the "source_id" for each event (for the same doi) and put them into one cell, delimited or seperated by a :::
This is the expression you use
forEach(value.parseJson().message.events,v,v.source_id).join(":::")
In my example below, all the events for that doi are Wikipedia related (different edits at different times or to different pages) so the output column is simply
wikipedia:::wikipedia:::wikipedia..... etc
Hopefully you are skilled enough this now makes sense.
Once you have extracted this, you can study if being mentioned or cited in Twitter, blogs, Reddit etc has any impact on citations.
Conclusion
There you have it. A beginner's guide to using APIs to pull in data for analysis. These aren't the only APIs of course, there are other APIs you can consider playing with. These include
Crossref API (citation data)
Elsever's Scopus and Sciencedirect API
The methods detailed above can work in a similar manner with most RESTFUL APIs, though some may require you to generate a APIKEY for you to add in the api query.
Still, most RESTFUL apis can be handled in a similar matter to what I have shown above. With APIs you can mix and match data in ways you couldn't easily in the past, so I hope this was useful.

